Writing Data To A Text File In Python
Overview When you’re working with Python, you don’t need to import a library in order to read and write files. It’s handled natively in the language, albeit in a unique manner.
A bit explanation: Your first question is, I want to remove duplicate IP? 7 wonders of the ancient world game. Here, you can use a set() on list, but then your requirement is to keep the unique items in. How do you append to a file? That a indicates the appending the file, that means allow to insert extra data to the. Writing to the end of a text file in python.
The first thing you’ll need to do is use Python’s built-in openfunction to get a file object. The openfunction opens a file.
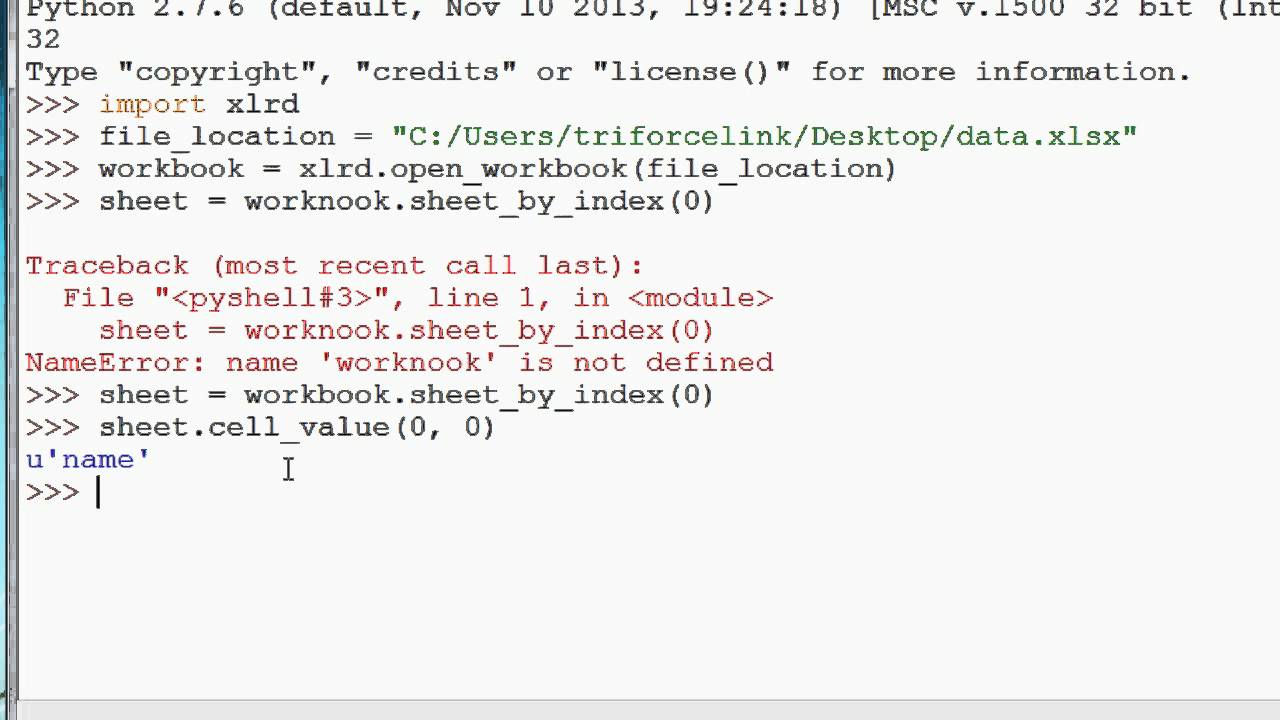
When you use the openfunction, it returns something called a file object. File objects contain methods and attributes that can be used to collect information about the file you opened. They can also be used to manipulate said file. For example, the modeattribute of a file object tells you which mode a file was opened in.
And the nameattribute tells you the name of the file that the file objecthas opened. You must understand that a fileand file objectare two wholly separate – yet related – things.
I want to redirect the print to a.txt file using python. I have a 'for' loop, which will 'print' the output for each of my.bam file while I want to redirect ALL these output to one file. So I tried to put f = open('output.txt','w'); sys.stdout = f at the beginning of my script. However I get nothing in the.txt file.
My script is: #!/usr/bin/python import os,sys import subprocess import glob from os import path f = open('output.txt','w') sys.stdout = f path= '/home/xug/nearline/bamfiles' bamfiles = glob.glob(path + '/.bam') for bamfile in bamfiles: filename = bamfile.split('/')-1 print 'Filename:', filename samtoolsin = subprocess.Popen('/share/bin/samtools/samtools','view',bamfile, stdout=subprocess.PIPE,bufsize=1) linelist= samtoolsin.stdout.readlines print 'Readlines finished!' So what's the problem? Any other way besides this sys.stdout? I need my result look like: Filename: ERR001268.bam Readlines finished! Mean: 233 SD: 10 Interval is: (213, 252). The most obvious way to do this would be to print to a file object: with open('out.txt', 'w') as f: print f, 'Filename:', filename # Python 2.x print('Filename:', filename, file=f) # Python 3.x However, redirecting stdout also works for me. It is probably fine for a one-off script such as this: import sys origstdout = sys.stdout f = open('out.txt', 'w') sys.stdout = f for i in range(2): print 'i = ', i sys.stdout = origstdout f.close What is the first filename in your script?
I don't see it initialized. My first guess is that glob doesn't find any bamfiles, and therefore the for loop doesn't run. Check that the folder exists, and print out bamfiles in your script. Also, use to manipulate paths and filenames. Or API reference: print(.objects, sep=' ', end=' n', file=sys.stdout, flush=False) The file argument must be an object with a write(string) method; if it is not present or None, will be used.
Since printed arguments are converted to text strings, print cannot be used with binary mode file objects. For these, use file.write(.) instead. Since normally contains write method, all you need to do is to pass a into its argument. Write/Overwrite to File with open('file.txt', 'w') as f: print('hello world', file=f) Write/Append to File with open('file.txt', 'a') as f: print('hello world', file=f). The easiest solution isn't through python; its through the shell.
From the first line of your file ( #!/usr/bin/python) I'm guessing you're on a UNIX system. Just use print statements like you normally would, and don't open the file at all in your script.
When you go to run the file, instead of./script.py to run the file, use./script.py where you replace with the name of the file you want the output to go in to. The token tells (most) shells to set stdout to the file described by the following token. One important thing that needs to be mentioned here is that 'script.py' needs to be made executable for./script.py to run.
So before running./script.py,execute this command chmod a+x script.py (make the script executable for all users). You may not like this answer, but I think it's the RIGHT one.
See More On Stackoverflow
Don't change your stdout destination unless it's absolutely necessary (maybe you're using a library that only outputs to stdout??? Clearly not the case here). I think as a good habit you should prepare your data ahead of time as a string, then open your file and write the whole thing at once.
This is because input/output operations are the longer you have a file handle open, the more likely an error is to occur with this file (file lock error, i/o error, etc). Just doing it all in one operation leaves no question for when it might have gone wrong. Here's an example: outlines = for bamfile in bamfiles: filename = bamfile.split('/')-1 outlines.append('Filename:%s'% filename) samtoolsin = subprocess.Popen('/share/bin/samtools/samtools','view',bamfile, stdout=subprocess.PIPE,bufsize=1) linelist= samtoolsin.stdout.readlines print 'Readlines finished!' Outlines.extend(linelist) outlines.append(' n') And then when you're all done collecting your 'data lines' one line per list item, you can join them with some ' n' characters to make the whole thing outputtable; maybe even wrap your output statement in a with block, for additional safety (will automatically close your output handle even if something goes wrong): outstring = ' n'.join(outlines) outfilename = 'myfile.txt' with open(outfilename, 'w') as outf: outf.write(outstring) print 'YAY MY STDOUT IS UNTAINTED!!!' However if you have lots of data to write, you could write it one piece at a time.
I don't think it's relevant to your application but here's the alternative: outfilename = 'myfile.txt' outf = open(outfilename, 'w') for bamfile in bamfiles: filename = bamfile.split('/')-1 outf.write('Filename:%s'% filename) samtoolsin = subprocess.Popen('/share/bin/samtools/samtools','view',bamfile, stdout=subprocess.PIPE,bufsize=1) mydata = samtoolsin.stdout.read outf.write(mydata) outf.close. @Gringo: I fail to see how my comment contradicts itself. Maybe the performance aspect isn't relevant, keeping a file handle open for an extended period always increases the risk of error.
In programming file i/o is always inherently more risky than doing something within your own program, because it means you have to reach out through the OS and mess around with file locks. The shorter you have a file open for, the better, simply because you don't control the file system from your code. Xrange is different because it has nothing to do with file i/o, and FYI I rarely use xrange either; cheers – Aug 22 '11 at 23:36. Don't use print, use logging You can change sys.stdout to point to a file, but this is a pretty clunky and inflexible way to handle this problem. Instead of using print, use the module. With logging, you can print just like you would to stdout, or you can also write the output to a file.
You can even use the different message levels ( critical, error, warning, info, debug) to, for example, only print major issues to the console, but still log minor code actions to a file. Changing the value of sys.stdout does change the destination of all calls to print. If you use an alternative way to change the destination of print, you will get the same result. Your bug is somewhere else:.
it could be in the code you removed for your question (where does filename come from for the call to open?). it could also be that you are not waiting for data to be flushed: if you print on a terminal, data is flushed after every new line, but if you print to a file, it's only flushed when the stdout buffer is full (4096 bytes on most systems).